Research
Automated Radiology Report Labelling
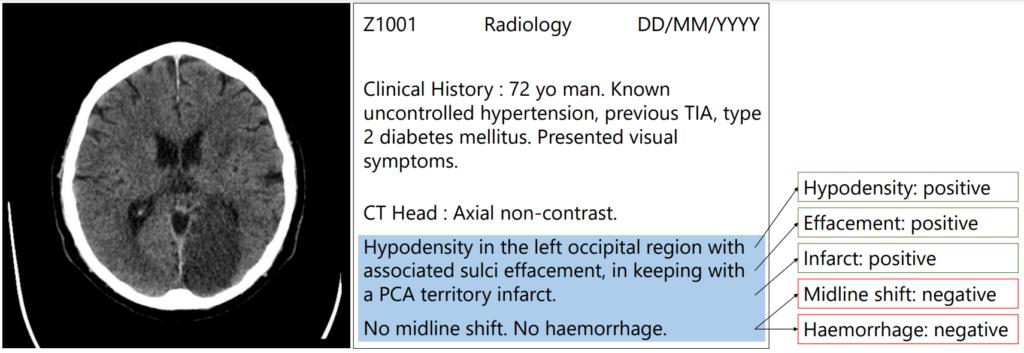
The goal of artificial intelligence and machine learning in healthcare is to help alleviate some of the pressure that is placed on doctors and help them make the right decisions more quickly and easily. Medical professionals often work long hours and have very busy schedules – they have many patients to see, many prescriptions to write and many diagnoses to make. When it comes to sifting through medical scans, radiologists spend most of their time looking at the “normal” scans rather than focusing their time on the scans that need their expert interpretation. Machine learning can help in these situations, however training machine learning algorithms requires large amounts of expertly annotated data which is time-consuming and expensive to obtain.
Patrick’s research focuses on how we can use medical text with small amounts of annotated data to provide accurate labels for large datasets that can be used to train image analysis algorithms. Medical scans are usually accompanied by free-text radiology reports which are a rich source of information. We automatically extract structured labels from head CT reports for imaging of stroke patients. Using novel approaches in deep learning and synthetic data augmentation, we are able to robustly extract labels classified according to the radiologist’s reporting. In a radiology report, each label can either be classed as a positive, uncertain or negative mention or it may not be mentioned at all. This approach can be used in further research to effectively extract many labels from medical text.
Generative Deep Learning in Digital Pathology

Diagnosing pathologies, like cancer, is highly skilled work involving experts looking at biopsy cross-sections slides under a microscope. In the last 5-10 years this process has partially digitised, with doctors swapping microscopes for high-resolution scanners and large displays.
What if we could generate synthetic data that could be used to train a supervised machine learning system? It would be anonymous and plentiful. This project is about trying to create synthetic pathology data using computer graphics and data refinement using generative deep learning. We are currently assessing several generated data sets and have some interesting initial results.
Smart homes for the elderly to promote health and well-being for independent living / Automated Remote Pulse Oximetry System
Pireh’s research focuses on smart home for the elderly to promote their health and wellbeing for independent living by monitoring their vitals to understand their health status and provide support when immediate attention is required. She is working on ARPOS (Automated Remote Pulse Oximeter System) which is to find an alternative way to help monitor the physical health of people, especially the elderly in a non-contact based manner while catering to the ethical and acceptance issues.
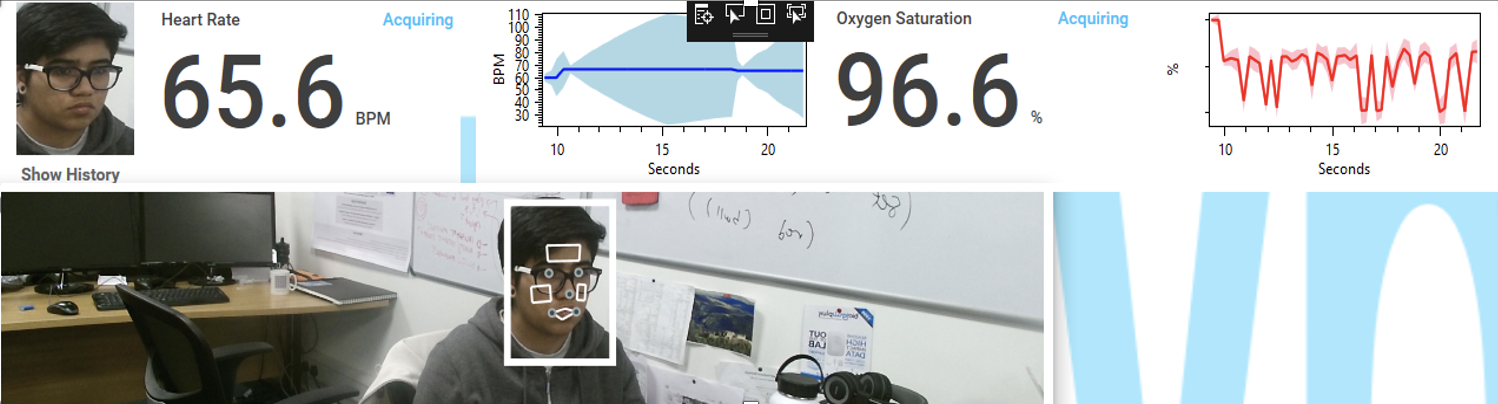
ARPOS uses non-contact automated remote sensing using camera-based technology to measure vital signs (heart rate, blood oxygen level and temperature) in real-time at a distance of up to 4.5m away from the person, measuring up to 6 people at once. This is also useful for other use cases such as the elderly with later stage dementia where monitoring vitals using contact-based sensors can be difficult, home, hospitals and psych wards.
Industrial Centre for AI Research in Digital Diagnostics
iCAIRD is a pan-Scotland Collaboration of 15 partner organisations including academics, NHS and industrial. It is centred at the University of Glasgow’s Clinical Innovation Zone at the Queen Elizabeth University Hospital. At the University of St Andrews, data scientists (Mahnaz Mohammadi, David Morrison and Christina Fell), are working with pathologists from NHSGG&C to develop & train AI on cases from the national database for automated reporting of endometrial and cervical biopsies. The developed algorithms will be integrated into clinical workflow, and using the national reporting system, can be scaled across the whole of NHS in Scotland with the aim of reducing gynaecological pathology reporting time by 50%. The key objectives of the project are:
- To create an AI algorithm for classification of endometrial neoplasia (adenocarcinoma, hyperplasia with atypia) from biopsies.
- To create an AI algorithm for identification of cervical biopsies as either benign or cancerous. For cancerous biopsies, classification of these as: invasive squamous or adeno-carcinoma, intra-epithelial neoplasia (low grade (including HPV and CIN1) and high grade (including CIN2 and CIN3).